研究小组认为,甲型H1N1流感病毒在2009年1月已经进入人体,但具体在哪个地域进入人体尚不清楚,可能在北美。而且,甲型H1N1流感病毒毒株是自然进化形成的,而非出自某个实验室。
此项研究还建议,要加强对猪群流感病毒的系统监测。
生物通网站刊登了对管轶的访谈。
1.State Key Laboratory of Emerging Infectious Diseases & Department of Microbiology, Li Ka Shing Faculty of Medicine, The University of Hong Kong, 21 Sassoon Road, Pokfulam, Hong Kong SAR, China
of Evolutionary Biology, University of Edinburgh, Ashworth Laboratories, King's Buildings, Edinburgh EH9 3JT, UK
3.Department of Ecology and Evolutionary Biology, University of Arizona, Tucson, Arizona 85705, USA
4. Department of Zoology, University of Oxford, South Parks Road, Oxford OX1 3PS, UK
Nature advance online publication 11 June 2009 | doi:10.1038/nature08182; Received 24 May 2009; Accepted 4 June 2009; Published online 11 June 2009
Correspondence to: Yi Guan1Andrew Rambaut2 Correspondence and requests for materials should be addressed to A.R. (Email: a.rambaut@) or Y.G. (Email: ).
【Abstract】
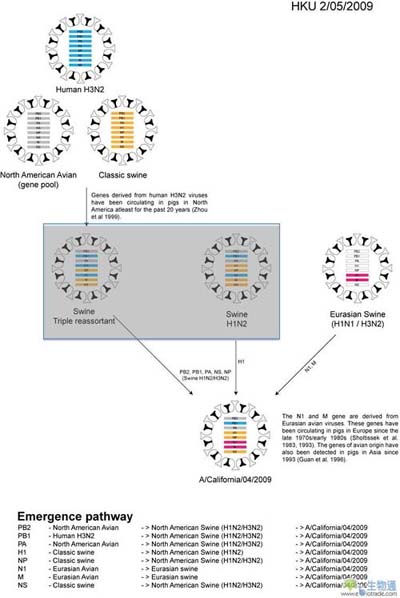
In March and early April 2009, a new swine-origin influenza A (H1N1) virus (S-OIV) emerged in Mexico and the United States1. During the first few weeks of surveillance, the virus spread worldwide to 30 countries (as of May 11) by human-to-human transmission, causing the World Heath Organisation to raise its pandemic alert to level 5 of 6. This virus has the potential to develop into the first influenza pandemic of the twenty-first century. Here we use evolutionary analysis to estimate the timescale of the origins and the early development of the S-OIV epidemic. We show that it was derived from several viruses circulating in swine, and that the initial transmission to humans occurred several months before recognition of the outbreak. A phylogenetic estimate of the gaps in genetic surveillance indicates a long period of unsampled ancestry before the S-OIV outbreak, suggesting that the reassortment of swine lineages may have occurred years before human emergence, and that the multiple genetic ancestry of S-OIV is not indicative of an artificial origin. Furthermore, the unsampled history of the epidemic means that the nature and location of the genetically closest swine viruses reveal little about the immediate origin of the epidemic, despite the fact that we included a panel of closely related and previously unpublished swine influenza isolates. Our results highlight the need for systematic surveillance of influenza in swine, and provide evidence that the mixing of new genetic elements in swine can result in the emergence of viruses with pandemic potential in humans.
0
推荐




 京公网安备 11010502034662号
京公网安备 11010502034662号 